DeepRepViz is an interactive web
tool that can be used to inspect the 3D representations learned by
predictive deep learning (DL) models. It can be used to inspect a trained
model for any biases in the training and the data and debug the model. The
tool is intended to help improve the transparency of such DL models. The
DL models can be a ‘black box’ and hinder their adaptation to critical
decision processes (ex: medical diagnosis). This tool aims to provide a
platform that developers of DL models can use to gain a better intuition
about what their model is learning by visualizing its ‘learned
representation’[1] of the
data. It can be used to understand what the model might be basing its
decisions upon.
With DeepRepViz, one can ask questions such as:
Developers can also share their model’s learned representation and the list of suspicious risk variables[2] that can be then independently inspected and verified by other stakeholders and users of the model.
As an example, consider the below (demonstrative) scenario:
Say, you developed a DL model that can diagnose Alzheimer’s disease from brain MRI with an accuracy of 80% on your test data. However, when a clinician tests your model in their hospital they complain that it is inaccurate and doesn’t perform better than chance (50%). Now you suspect that something must have gone wrong in your DL model training process or in the data used to train the model that made it incapable of generalizing to the subjects in the hospital. Either your model learned to predict Alzeihmers using some spurious shortcuts. Maybe the age of the subjects correlated with the label such that the healthy controls in your data were always younger than Alzheimer’s subjects. Or the MRI scanner machines used were different for Alzheimer’s patients and healthy controls as they were scanned at different locations. Or there is a complex spurious interaction effect in the data (as happens to be in reality) such that older subjects and subjects scanned on scanner A by chance seemed to 4 times more likely to be Alzheimer’s patients and your DL model just picked up on this relation to get 80% accuracy.
To verify, you prepare a list of ‘suspicious’ variables from your dataset, extract your model’s learned representation, upload it on DeepRepViz and inspect them (as shown in section How does DeepRepViz work?.
After testing all your suspicious variables you find that the variableage X scannerindeed shows a significantly high ‘confounding risk score’. You retrain your model by resampling the data such that this spurious relation between theage X scannerand the diagnosis label is removed. When you test the new model’s representation on the tool you see that none of the suspicious variables, includingage X scannerhave a high confounding risk score anymore.
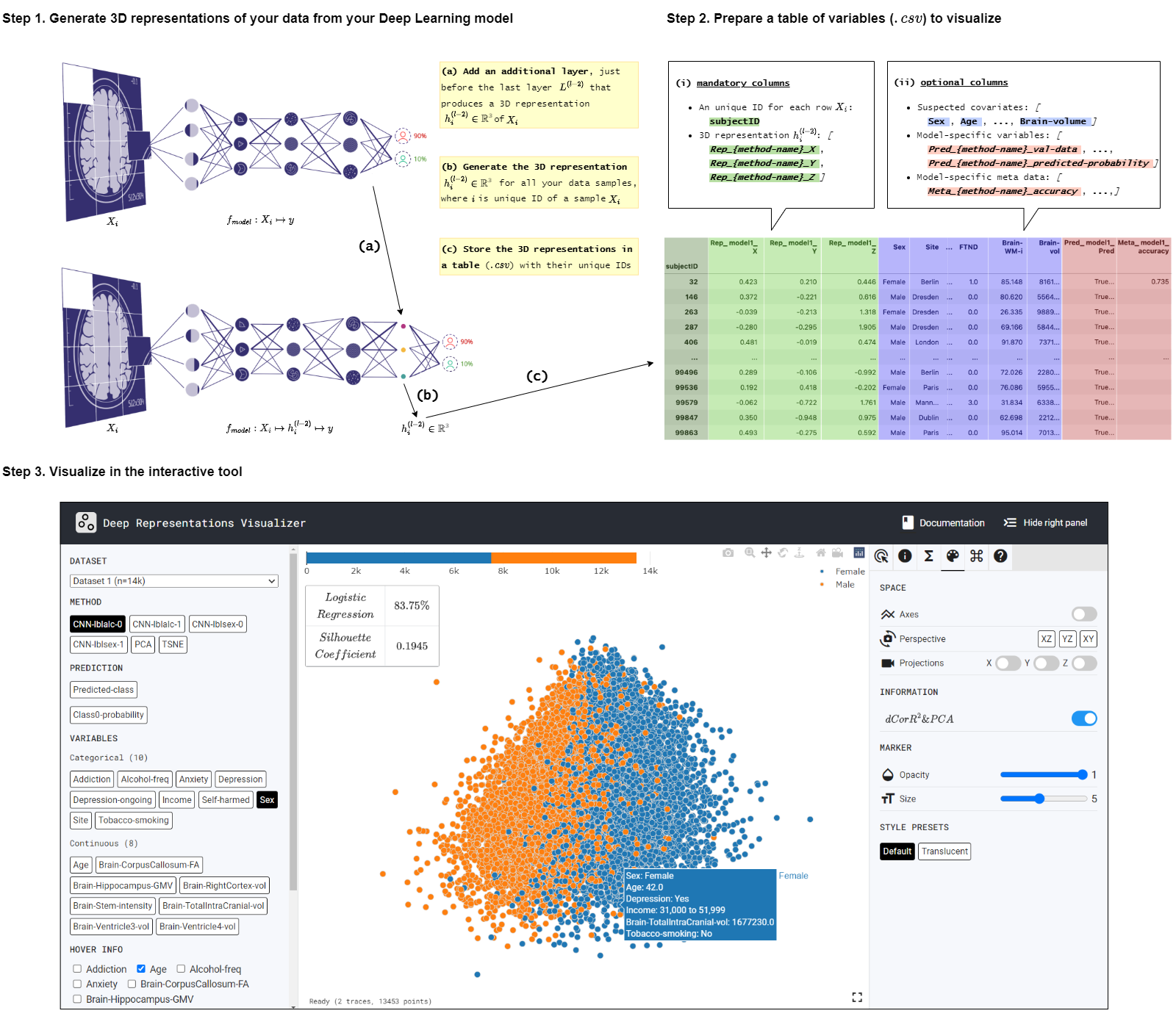
Consider a DL model fmodel : X ↦ y with with l layers {L(0), L(1), ....L(l−2), L(l−1)}. To inspect this model using DeepRepViz the next 3 steps can be followed (An intuitive illustration of the procedure is provided below):

Add an fully-connected layer with 3 weights and no bias parameter, just before the final layer such that fmodel : X ↦ h(l−2) ↦ y where h(l−2) ∈ ℝ3 and the model has l layers. This layer forces the DL model to learn an intermediary 3-dimensional (3D) representation that we can visualize and inspect in the DeepRepViz tool.
To save this 3D representation:
.csv) to visualize
The csv file uploaded on the tool will work only if it contains the
below columns and column name formating:
subjectID (N=1): Unique IDs for each row. Acts
as an index for the table. It can be any user-defined unique name
used to identify a specific model and prediction task it performs.
As an example, if users are trained a Convolutional Neural Network
(CNN) model to predict their age from brain MRI then the columns can
be named as [Rep_CNN-brainage_X],
[Rep_CNN-brainage_Y], and
[Rep_CNN-brainage_Z].
Rep_[method name]_X, Rep_[method name]_Y,
Rep_[method name]_Z] (N=1 x 3): The
[X,Y,Z]
coordinates of the generated 3D representation space
hi(l−2) ∀i ∈ {0..N − 1}. These coordinates determine the position of each subject along
with a unique ID subjectID in the 3D space of the viz
tool.
[variable name](optional) (N=inf):
These are the variables whose distribution you wish to visualize
with respect to the 3D representation space. When this variable is
selected on the tool, each subject is colored by the value of this
variable. Append all potential confounding variables to the table as
additional columns. Variables with numerical values will be
interpreted by the tool as an ordinal or an interval type and
variables with string values will be interpreted as nominal type.
However, this can be interchanged later in the tool. There is no
limit to how many variables are added. In the above example these
are the variables age, sex,
FTND, total brain volume,
ventricles size and hippocampus area. Can
be continuous or categorical.
Pred_[method name]_[pred info name] (optional)
(N=inf): You can provide method-specific information for
each method about the each subject. Can be continuous or
categorical. Ex: for methods using machine learning / deep learning
models, you can provide which subjects were predicted correctly,
which subjects were used as the test subjects, what was the
probability of model prediction for each subject.
Meta_[method name]_[meta info name] (optional)
(N=inf): You can provide method-specific meta data
information that you want to show in the tool in general. Ex: for
methods using machine learning / deep learning models, you can
provide the prediction accuracy of that model and information about
the model architecture. Ex2: for PCA method you can provide the % of
explained variance by
[X,Y,Z].(Note 1: text under [] such as [method name] and [variable name] means it can be anything the user chooses and it will be displayed by that name in the tool)
(Note 2: When the columns marked as (optional) are not included in the csv, the tool will still work.)
(Note 3: (N=i) indicates how many of these column instances are expected. (N=inf) implies that there is no limit to the number of these columns)
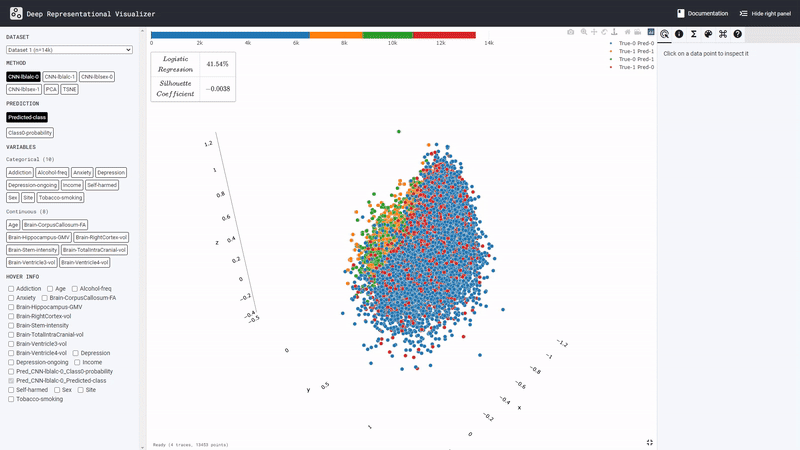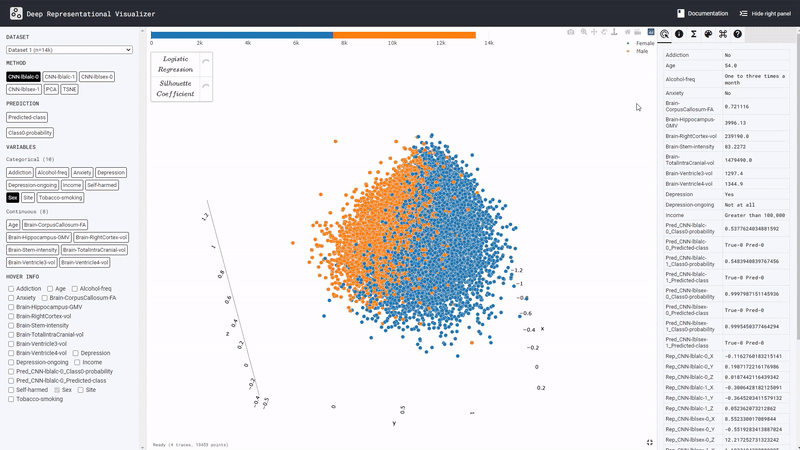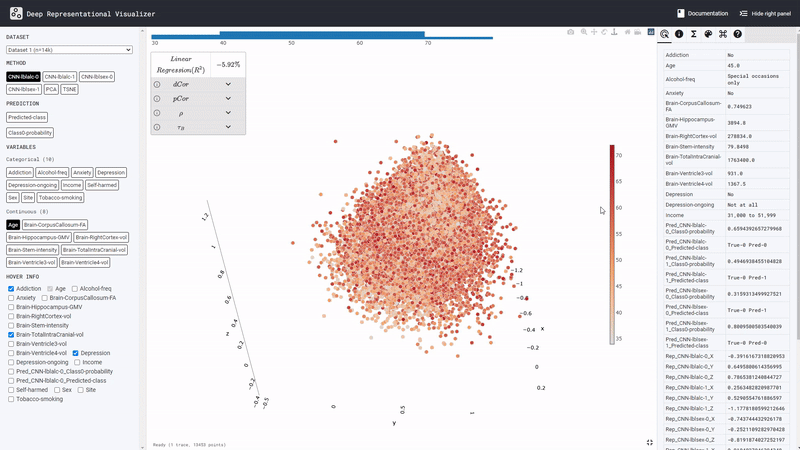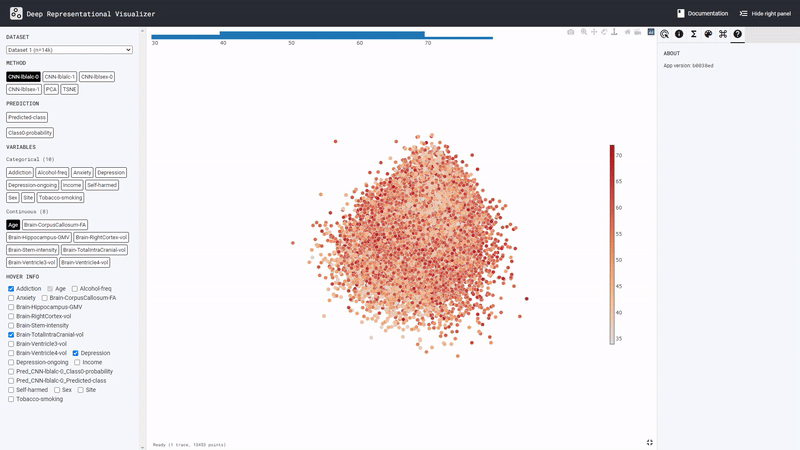
The learned representation space of the DL model is displayed at the center. Users can interactively inspect this representation space using various features available in the panel on the right. From the panel on the left, users can select different ‘potential confound’ variables and color-code the representation space in the center. The tool also computes the confounding risk scores for the selected variable as shown in the top left corner of the central display. Find out more in the next section Graphic User Interface (GUI) Features.
Let’s take a closer look at the components of the
DeepRepViz interface:
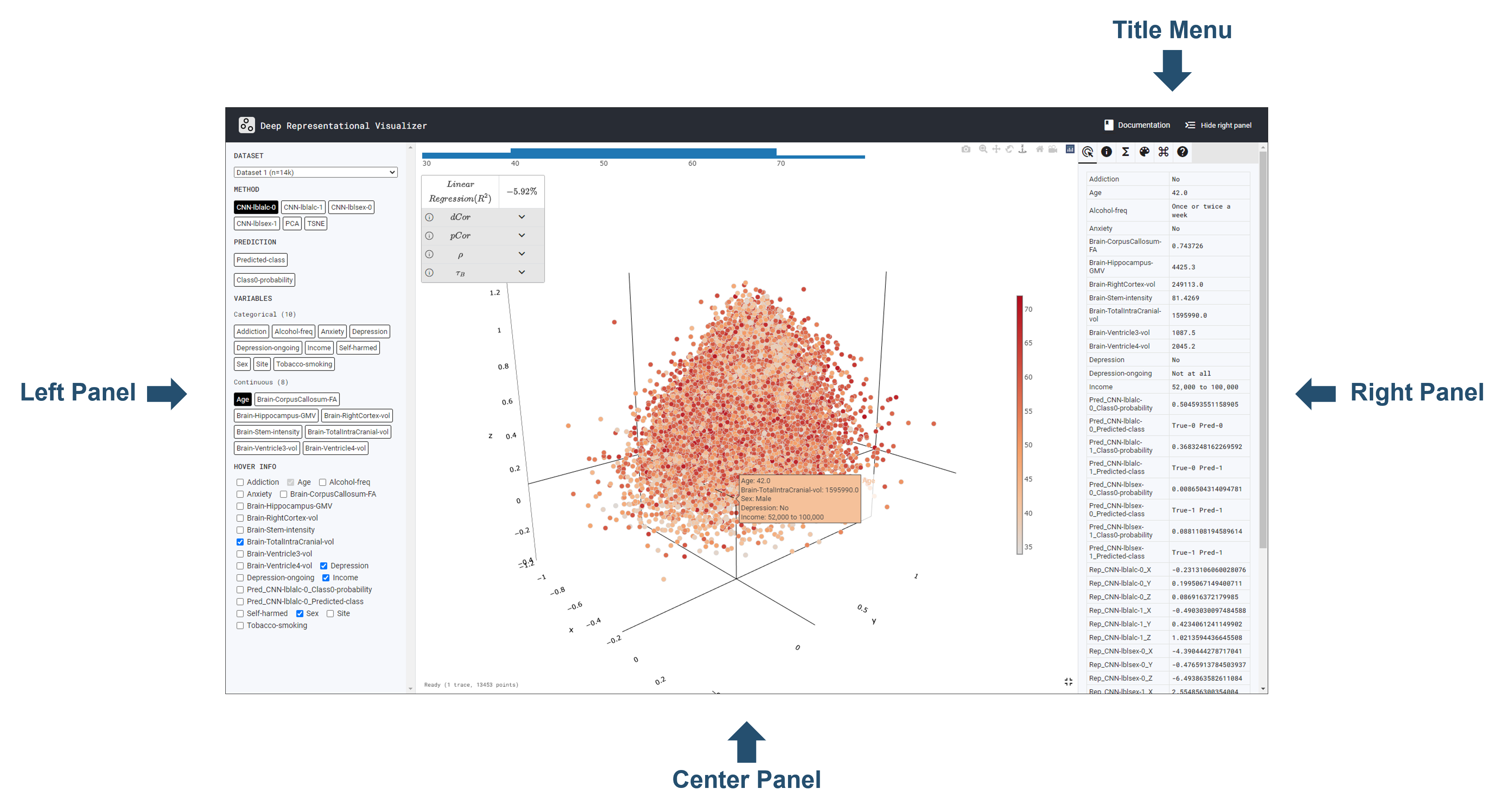
 Documentation hyperlink to the
DeepRepViz documentation,
and
Documentation hyperlink to the
DeepRepViz documentation,
and
 Hide Right Panel, this function hides the right
panel.
Hide Right Panel, this function hides the right
panel.
The left panel has four areas; Dataset,
Method, Colum, and
Hover Info from top to bottom.
.csv) to
visualize. Full Screen supports full screen function.
Full Screen supports full screen function.
The
 Settings Bar under the
Title Menu supports the following functions for
representation space:
Settings Bar under the
Title Menu supports the following functions for
representation space:
 Datapoint represent each columns information.
Datapoint represent each columns information.
 Metadata only if shows the model’s metadata; for
examples, please check the dataset 2 case.
Metadata only if shows the model’s metadata; for
examples, please check the dataset 2 case.
 Transform can do such as Min-Max Scaling,
Standardization, PCA and displayed ALL Stats.
Transform can do such as Min-Max Scaling,
Standardization, PCA and displayed ALL Stats.
 Styling users can do such space, infromation, and
maerker change. It also provided style presets.
Styling users can do such space, infromation, and
maerker change. It also provided style presets.
 Shortcuts, it provides how to use the shorcuts
key.
Shortcuts, it provides how to use the shorcuts
key.
 About shows the App version.
About shows the App version.
In the below examples, inspecting two predictive DL models using
DeepRepViz helped us find
spurious confounder variables and biases in the models:
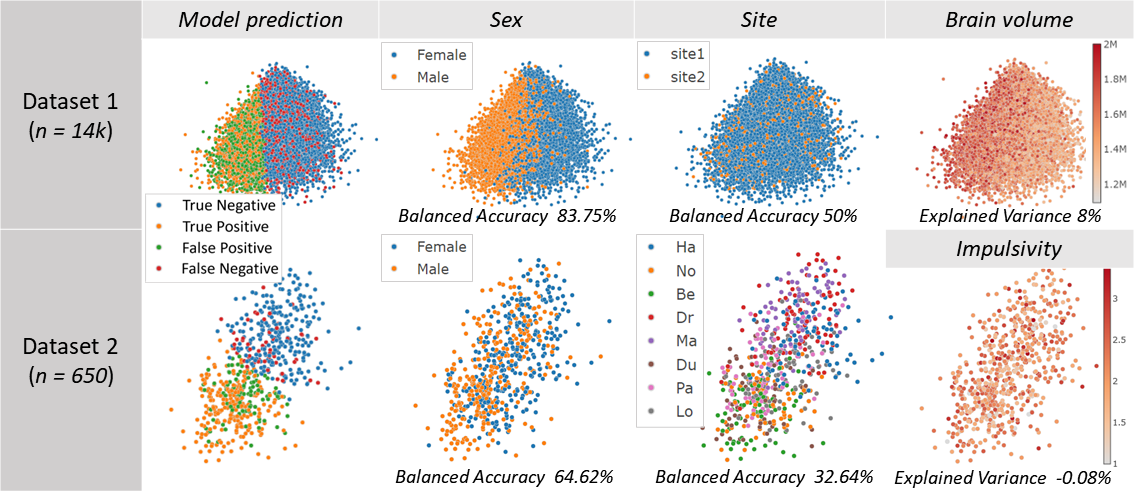
We predicted alcohol drinking frequency from brain MRI. Model prediction boundary aligns withSexandBrain volumeof the subjects, but not with theSitevariable. The confounding risk scores also reflects this. This suggests that the DL model may have simply predicted all males in the data and subjects with large brain volume as frequent drinkers.
We predict a social behavior (detail held for anonymity) from brain MRI. Here theSitevariable seems to confound the model predictions. It can be observed, for instance, that all subjects from the Dr site (see legend ofSite) are categorized by the model as having risky category of the social behavior, whereas all subjects from the No site are exclusively predicted as have a safe social behavior. To prevent this spurious effect, the MRI dataset can be harmonized across the different sites where the data was collected.
This project was inspired by Google Brain’s projector.tensorflow.org, but is more catering towards the medical domain and medical imaging analysis. For implementation, we heavily rely on 3D-scatter-plot from plotly.js.
[1] Bengio, Yoshua, Aaron Courville, and Pascal Vincent. “Representation
learning: A review and new perspectives.” IEEE transactions on pattern
analysis and machine intelligence 35.8 (2013): 1798-1828.
[2] Görgen, Kai, et al. “The same analysis approach: Practical protection
against the pitfalls of novel neuroimaging analysis methods.” Neuroimage
180 (2018): 19-30.
Last updated 2022-10-24 UTC.